We study molecular evolution, systematics, phylogeography, comparative genomics and behavioral evolution of birds and (occasionally) non-avian reptiles.
Natural history drives our research

Many of our research questions are motivated by natural history observations. This applies to our more ecologically-oriented work, such as undergraduate Elaine Vo's investigation of organic mercury levels in black footed albatrosses (Vo et al. 2011), as well as to our more molecular work. Genomics and studies of genome evolution provide a remarkable window into the molecular basis of interesting phenotypic variation. Scott has had a long-term interest in seabirds of the order Procellariformes, which includes the albatrosses and petrels. These birds, with their long-distance foraging journey and remarkable life histories, are not only inspiring but also offer a treasure-trove of interesting molecular ecology facts to discover. Our recent work on procellariformes includes the organic mercury study, as well as population genetic studies of black-footed albatrosses, and finally our ongoing work with Gaby Nevitt (UC Davis) on chemical ecology and mate recognition in Leach's Storm Petrels.
Comparative genomics in birds and other reptiles
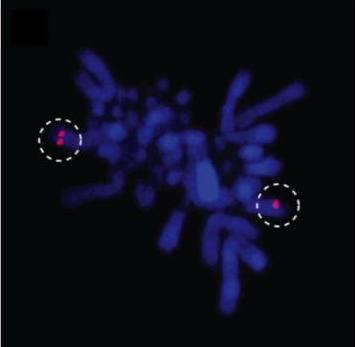 This picture shows the karyotype of an emu, a flightless bird now restricted to the arid regions of Australia. The red dots in the photo indicate the locations of specific genes, as revealed by FISH (fluoresecent in-situ hybridization) experiments using individual bacterial artificial chromosome (BAC) clones. Our work on Emus, published in Journal of Heredity in collaboration with colleagues from Australia, exemplifies our use of comparative genomics resources that few others have access to, including BAC libraries from several birds and reptiles, made in collaboration with Chris Amemiya of the Benaroya Research Institute in Seattle. These libraries were initially useful for gaining insight into the genomic architecture of various basal birds and reptiles (see Shedlock et al. 2007; Janes et al. 2008; Chapus et al. 2010), and are now proving useful as a supplement to improve genome assemblies in whole-genome sequencing projects. These BAC libraries have been used in several ongoing or completed genome projects nationally, including those for the Painted Turtle, American Alligator, Tuatara, Garter Snake, and Emu. Our ongoing whole-genome sequencing in House Finches is also benefitting from the fosmid library of a House Finch that we made in the 1990s, and which has served as a foundation for several papers (e.g., Hess et al. 2000). This work has expanded into comparative genomics studies using bioinformatics, such as our work examining isochors in Anolis (Fujita et al. 2011) or large conserved noncoding sequences (LCNSs) in amniotes (Janes et al. 2011).
This picture shows the karyotype of an emu, a flightless bird now restricted to the arid regions of Australia. The red dots in the photo indicate the locations of specific genes, as revealed by FISH (fluoresecent in-situ hybridization) experiments using individual bacterial artificial chromosome (BAC) clones. Our work on Emus, published in Journal of Heredity in collaboration with colleagues from Australia, exemplifies our use of comparative genomics resources that few others have access to, including BAC libraries from several birds and reptiles, made in collaboration with Chris Amemiya of the Benaroya Research Institute in Seattle. These libraries were initially useful for gaining insight into the genomic architecture of various basal birds and reptiles (see Shedlock et al. 2007; Janes et al. 2008; Chapus et al. 2010), and are now proving useful as a supplement to improve genome assemblies in whole-genome sequencing projects. These BAC libraries have been used in several ongoing or completed genome projects nationally, including those for the Painted Turtle, American Alligator, Tuatara, Garter Snake, and Emu. Our ongoing whole-genome sequencing in House Finches is also benefitting from the fosmid library of a House Finch that we made in the 1990s, and which has served as a foundation for several papers (e.g., Hess et al. 2000). This work has expanded into comparative genomics studies using bioinformatics, such as our work examining isochors in Anolis (Fujita et al. 2011) or large conserved noncoding sequences (LCNSs) in amniotes (Janes et al. 2011).
Chemical ecology and the molecular genetics of mate choice in Leach's Storm Petrels

In collaboration with Gabrielle Nevitt of UC Davis, we (including postdoc Simon Sin) have been carrying out a multi-year study on a colony of Leach's Storm Petrels in Nova Scotia, Canada. These birds are notable for their long lives, intensive parental investment, and impressive olfactory abilities. This combination of traits makes the storm petrel an ideal study species for understanding the role of the MHC (major histocompatibility complex) in avain mate choice. Existing research has demonstrated that certain organisms may use olfaction to discern differences in the MHC of possible mates, allowing for disassortative mating and greater immune response range of offspring. Further research and analysis will provide more insight into the role of the MHC gene family in petrel mate choice.
Ecological genomics of the evolution of pathogen resistance
In collaboration with Geoff Hill of Auburn University, we have been studying the evolutionary genomics of host-pathogen interactions, using the House Finch- Mycoplasma gallisepticum (MG) system as a model. This system is remarkable in several aspects. MG is usually found in poultry but in 1994 field scientists detected it in populations of House Finches introduced in the 1940s to the eastern US. The disease spread rapidly throughout house finches in the eastern US and is now found in all US states except a few in the southwest. Using a combination of genomic comparisons across geography, historical insights of museum specimens and experimental infections, we are reconstructing the history of spread and co-evolution between host and pathogen. Our recent work has involved gene expression analyses using microarrays (Bonneaud et al. 2011, 2012), whole genome sequencing of multiple Mycoplasma isolates using next-generation approaches (Delaney et al. 2012), and we are currently conducting candidate gene studies and whole genome sequencing to identify loci undergoing selective sweeps as a result of the epizootic. Our studies suggests that the House Finch-MG interaction suggest rapid change in both host and parasite, and that a third actor, namely viruses, may also be important for explaining the evolutionary dynamics of this system.
Phylogeography and historical demography of avian populations
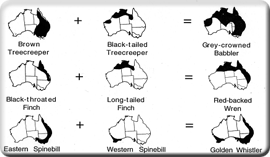
Our lab has a long tradition of studies phylogeography and systemtatics, extending back to Scott's dissertation research on Australian babblers. After twenty years of fieldwork in Australia, we have a large backlog of samples available for student research in phylogeography. We have also recently conducted phylogeographic surveys of several North American species inhabiting pine-oak woodlands, in collaboration with Garth Spellman of Black Hill State University in South Dakota. Our phylogeographic work is now complemented by climate models such as those produced by former postdoc Susan Cameron Devitt (see Edwards et al. 2012). Our work in phylogeography and systematics has championed the use of multiple loci to improve resolution and test various demographic hypotheses. Having started with mtDNA like everyone else (e.g., Edwards 1993), we quickly expanded to multiple loci isolated through genomic cloning (Jennings et al. 2005; Lee et al. 2008) and are now moving into the era of next generation sequencing. Allison Shultz, a graduate student in the lab, is now using the new rad-tag method to generate a large multilocus, multipopulation data set for the House Finch. Geographic variation is at the core of our work on virtually any problem; we envision many more such phylgeographic studies in the years to come.
Phylogenetics using the multispecies coalescent model
 Our lab has developed several methods for estimating phylogenetic trees using the so-called multispecies coalescent model (Liu et al. 2009a), and has championed their use (Edwards 2009). Much of the original theoretical work was conducted by a former postdoc, Liang Liu, who is now a professor in the Department of Statistics at the University of Georgia. The increase in scale of phylogenomic data sets certainly increases resolution of phylogenetic problems, but it also raises a number of analytical issues that are poorly dealt with by existing paradigms, such as the paradigm of combining information from multiple genes via supermatrices or concatenation.
Our lab has developed several methods for estimating phylogenetic trees using the so-called multispecies coalescent model (Liu et al. 2009a), and has championed their use (Edwards 2009). Much of the original theoretical work was conducted by a former postdoc, Liang Liu, who is now a professor in the Department of Statistics at the University of Georgia. The increase in scale of phylogenomic data sets certainly increases resolution of phylogenetic problems, but it also raises a number of analytical issues that are poorly dealt with by existing paradigms, such as the paradigm of combining information from multiple genes via supermatrices or concatenation.
Our recent demonstration that coalescent models outperform, and are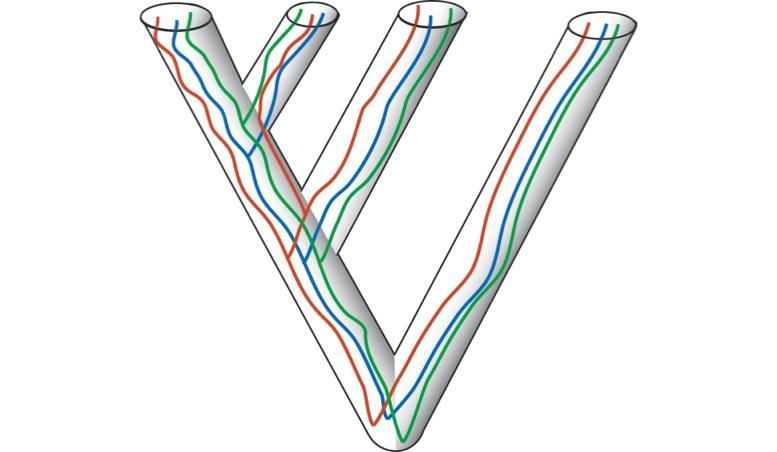 more consistent than, concatenation methods (Song et al. 2012) is now being applied to other phylogenomic data sets, such as birds. These are exciting times for systematics and we welcome students with interest in trees as tools to explore evolutionary processes, from genomics to patterns of organismal diversification.
more consistent than, concatenation methods (Song et al. 2012) is now being applied to other phylogenomic data sets, such as birds. These are exciting times for systematics and we welcome students with interest in trees as tools to explore evolutionary processes, from genomics to patterns of organismal diversification.

